Current & Past Projects

At CBHDS, we engage in a wide range of bioinformatics projects through active research collaborations and student mentoring. Our work spans genomics, transcriptomics, and epigenomics analyses, including customized database construction, non-model species genome annotation, orthology-based comparative analysis, RNA-Seq data analysis, functional pathway analysis, and DNA methylation profiling. We also conduct multimodal integrative studies that combine sequence, imaging, and phenotypic data to address complex biological questions. In addition to research, we are deeply committed to community engagement and education, developing tutorials, leading workshops, and providing hands-on training to help students and researchers build practical bioinformatics and data analysis skills. Through these efforts, we foster collaboration, enhance reproducibility, and expand access to computational biology tools and methods.
Since Fall 2022, we have collaborated with over 20 Virginia Tech researchers and students from the Departments of Biological Systems Engineering, Biological Sciences, Biochemistry, Biomedical Sciences and Pathobiology, and Human Nutrition, Foods, and Exercise, as well as the School of Plant and Environmental Sciences and the School of Medicine on 17 bioinformatics projects. These projects have included identifying biomarkers for pathogen and disease detection, annotating non-model species, identifying rare SNPs associated with specific patient genotypes, examining the roles of disease-defense genes, investigating treatment effects on different cell types, and studying the effects of tumor-suppressing genes and proteins in specific cell types.
- Sequence Quality Check
- Genome Assembly
- Custom Database Building:
- Custom protein/cDNA databases for annotation
- Custom repeat libraries
- Custom BLAST databases
- Functional databases integration (Combine GO, KEGG, InterPro, and Pfam terms for local use)
- Genome Annotation (especially for non-model species)
- Orthology and Comparative Genomics
- Ortholog/paralog identification
- Gene family clustering
- Pan-genome analysis
- Phylogenomics
- SNP and Variant Analysis
- Bulk & Single-cell RNA-seq
- Spatial Transcriptomics
- Differential Expression
- Functional Analysis
- Isoform and Splicing Analysis
- DNA Methylation Analysis
- Histone Modification and Chromatin Profiling
- ChIP-seq
- CUT&Tag
- CUT&RUN
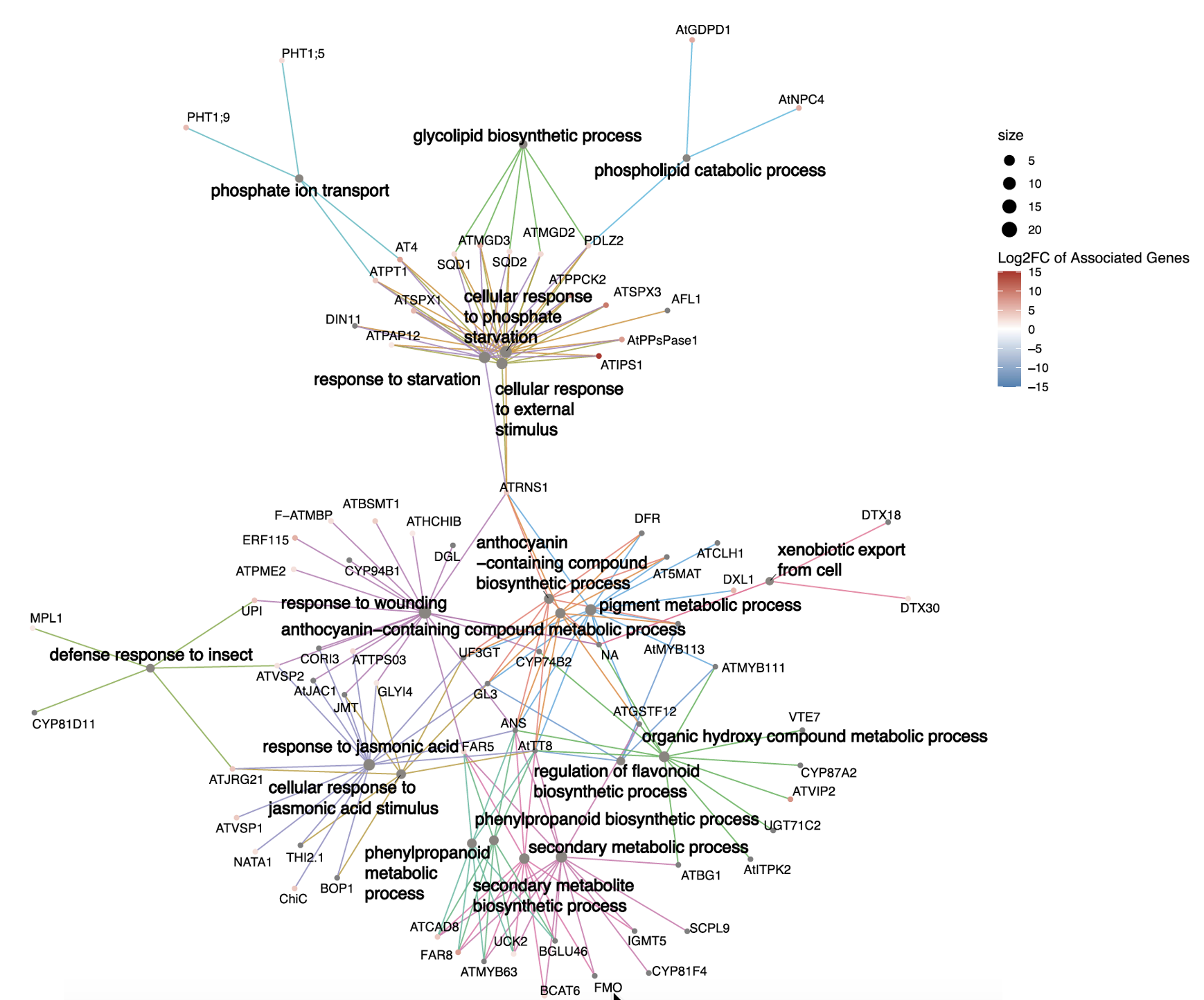
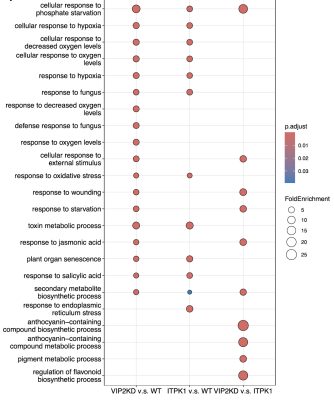
Cridland, C., Russo, A., Craige, B., Donahue, J., Zhang, X.M., Payne, M., Gillaspy, G. and Freed, C. (2025), Enhancing inositol pyrophosphate accumulation in plants alters growth, phosphate homeostasis, and insect herbivory. Plant J, 123: e70315. https://doi.org/10.1111/tpj.70315

Don’t see the analysis or resource you’re looking for? Contact us! We may still be able to help.
We provide training and research opportunities for students. We have mentored two Computational Modeling and Data Analytics (CMDA) student teams on their capstone projects, offering training in biological concepts, Python, R, UNIX commands, and project management. We also provide students and researchers with bioinformatics support during our weekly virtual drop-in sessions and we have been providing customized bioinformatics training to graduate students with no programming background. Please come to our drop-ins or send us a collaboration request if you are interested.
"For biologists and neuroscientists like me—people who come from wet labs and suddenly have to handle massive datasets—CBHDS is what makes that transition possible."
-Caroline de Jager, PhD, Translational Biology, Medicine, and Health.
Read Caroline's story
Our community engagement efforts include implementing a university-wide bioinformatics needs survey and preparing guidance documents to promote reproducible data analysis workflows, data management, and data security.
Our bioinformatics division is funded by the Fralin Life Sciences Institute at Virginia Tech to provide bioinformatics support to FLSI and external researchers and students. We are working closely with FLSI core facilities to facilitate research and publication for researchers within FLSI and beyond. These facilities include:
- Genomics Sequencing Center (GSC): a dedicated, multi-user resource for Next-Generation Sequencing (NGS) technologies offering rapid, cost-effective project planning as well as genomic, transcriptomic, and functional-genomics services;
- Facility for Advanced Imaging and Microscopy (FAIM): a multi-user resource providing state-of-the-art light microscopy instrumentation and expertise.
We collaborate with them to provide integrated support including data transfer and management, data analysis, and visualization, ensuring seamless support for researchers. Please contact us if you’re interested in integrated sequencing and imaging data collection and analysis support.