Student Projects
CMDA Student Capstone Projects
In the Computational Modeling and Data Analytics (CMDA) Capstone Project course at Virginia Tech, teams of three or four students spend the semester tackling an open-ended, client-driven project. In addition to the technical aspects of the project, students are mentored by CMDA Faculty in teamwork, project management, and technical leadership. Through the lens of their particular projects, the teams also consider the ethical aspects of data science and mathematical modeling.
2025 Projects
- Guiding Question: Can we develop a web application to consolidate and present essential information on graduate programs in statistics, biostatistics, data science, and related fields?
- Methods/tools: Web scraping, relational PostgreSQL database, Flask-based API
- Deliverable: An interactive web application with graduate program live filtering, individual program search bar, and head-to-head program comparisons
Team's Story
This project aimed to transform the American Statistical Association’s graduate program discovery platform from a manually maintained system into a fully automated, scalable data pipeline that helps students identify and compare statistics, biostatistics, and data science programs. Our team implemented automated web scraping, data validation routines achieving over 95% accuracy, a relational PostgreSQL database, and a Flask-based API to reliably collect, store, and serve program information. Analytic methods focused on validation of accuracy benchmarking, scrape performance evaluation, and database query efficiency testing. Visualizations included system architecture diagrams, database integration diagrams, and frontend interface views that demonstrate filtering, searching, and program comparison functionality for end users.

Experiential Narrative
Working on this project was a very collaborative, hands-on experience that felt closer to a real-world engineering team than a typical class assignment. As a team, we had to constantly communicate across different parts of the system: scraping, validation, database design, backend APIs, frontend development, and deployment, which meant no one could work in isolation for long. We ran into plenty of challenges along the way, especially when scrapers broke due to website structure changes or when integrating the database with the API and frontend, but solving those problems together was one of the most rewarding parts of the project. Everyone brought a different strength to the table, and over time we learned how to trust each other’s work, divide tasks more efficiently, and adapt when things didn’t go as planned. Overall, the project felt like building something that actually mattered, and it gave us valuable experience working as a team on a complex, end to end system rather than just individual components.
Project Team Members:
- Varun Kadari; B.S in Computational Modeling and Data Analytics (Minors in Computer Science and Mathematics); December 2025
- Jason Bruno Terceros; B.S in Computational Modeling and Data Analytics (Minor in Computer Science); December 2025
- Nate Montgomery; B.S in Computational Modeling and Data Analytics; May 2026
CBHDS Sponsors:
- Guiding Question: Can we develop a web application to consolidate and present essential information on graduate programs in statistics, biostatistics, data science, and related fields?
- Methods/tools: RSelenium and RShiny
- Deliverable: An interactive web application with graduate program live filtering, individual program search bar, and head-to-head program comparisons

Team's Story
The goal of the Program Discovery project, initiated by the American Statistical Association (ASA), was to develop a centralized, user-friendly web application that helps prospective graduate students explore and compare programs in statistics, biostatistics, and data science. The team used a hybrid data collection strategy—combining RSelenium-based scraping and manual verification—to compile detailed program information such as GPA requirements, GRE policies, tuition, and faculty expertise. The data was displayed through an interactive R Shiny dashboard featuring dynamic filters, a real-time keyword search bar, and side-by-side program comparison using modal pop-ups. These visual and interactive elements were designed to streamline the search process and support more informed decision-making for students.
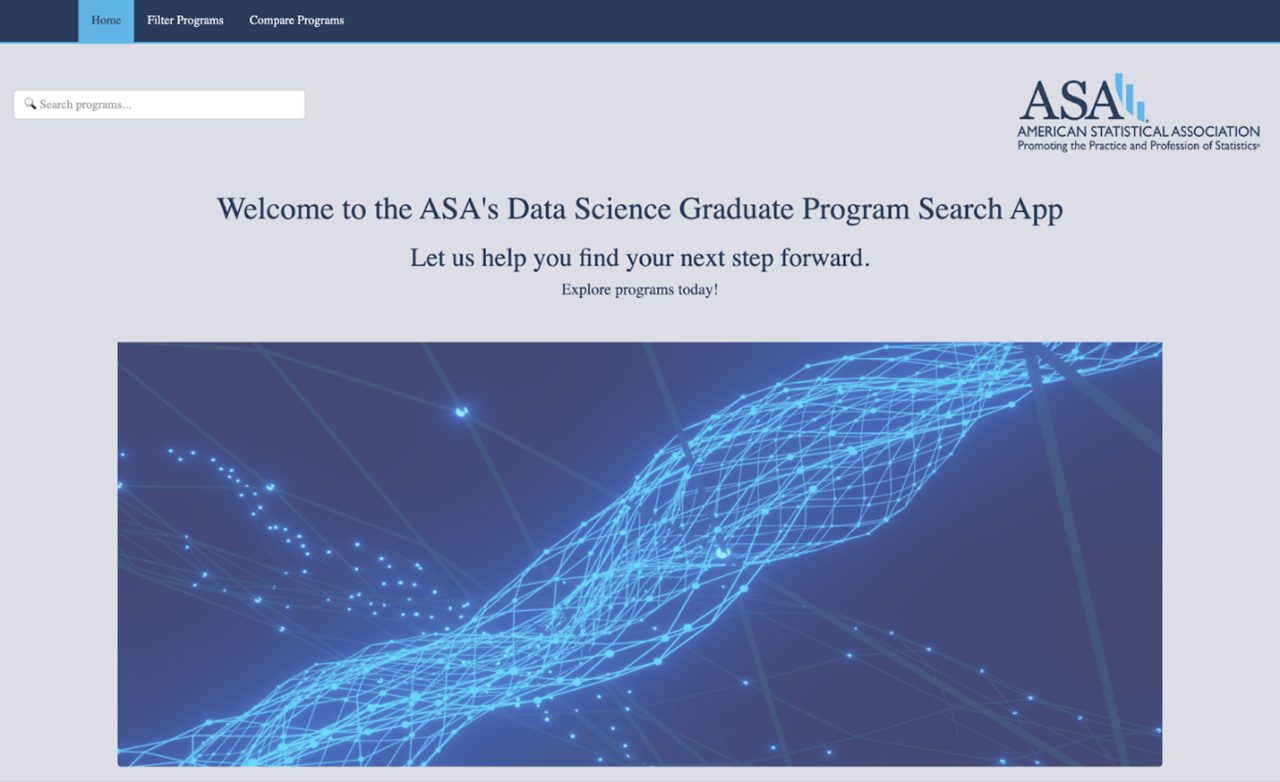
Experiential Narrative
Working on the Program Discovery project was one of the most hands-on and detail-oriented projects we’ve had during our time at VT. From the start, we realized just how messy and inconsistent university websites can be—and that collecting accurate data wasn’t as simple as running a script. We spent countless hours digging through program pages, course catalogs, and hidden PDFs just to find specific details like faculty expertise and GPA/GRE requirements. There were definitely moments of frustration (especially when websites refused to load or buried key info five clicks deep), but we also got into a rhythm and learned how to divide and conquer. Building the app itself was a creative process—figuring out how to make the user interface intuitive while keeping the backend clean and functional. We learned a lot about collaboration, project planning, and how to troubleshoot problems in real time. By the end, it was really satisfying to see everything come together into something that could genuinely help students like us.
Project Team Members:
- Carly Bransford, Computational Modeling and Data Analytics, Virginia Tech Class of ‘25.
- Meghna Varali Kommoju, Computational Modeling and Data Analytics, Virginia Tech Class of ‘25.
- Hyeonmin Kim, Computational Modeling and Data Analytics, Virginia Tech Class of ‘25.
CBHDS Sponsors:
- Guiding Questions:
Could the FDA have detected a disproportionate association between stroke-related adverse events and Lemtrada earlier using the FAERS database?
Do other multiple sclerosis drugs also show disproportionate associations with stroke-related adverse events? - Analytical methods: Time-series analysis, Disproportionality Analysis, Logistic Regression
- Visual Analytics: Time-series line charts

Team's Story
Our study evaluated six FDA-approved MS drugs using a combination of disproportionality and time-series analyses to determine whether FAERS data could reveal early indications of stroke-drug interaction signals that might warrant regulatory action. Among the six drugs analyzed, only Lemtrada and Rebif displayed a significant disproportional relationship with stroke adverse events. While Lemtrada has had known stroke risk since 2018, this is the first safety signal reported for Rebif. Notably, our findings identify a safety signal for Lemtrada onset almost two years prior to the FDA boxed warning. Rebif has displayed a similar signal since 2010, with no current warnings in place. Overall, our findings show that signal detection methods in FAERS can reveal early drug safety risks which could support faster FDA intervention.


Experiential Narrative
At first when our team started the project, we felt overwhelmed by the immensity of the project’s goal and the gravity of the scope. Though some of us had experience working with healthcare data in the past, this deep dive into multiple sclerosis and its adverse effects was very new to us. All of us are very accustomed to R and have worked with it greatly in the past, however the methods used in our project were well out of our comfort zone. The work felt gradual and continuous however, when we could start to see progress and especially identifying Rebif as a potential candidate for a black box warning, it felt very rewarding. Working with so many different analytical methods taught us a certain diligence to data and made us highly questionable of initial results, but with the help of our sponsors' navigation we became more and more confident throughout the project. We are especially grateful to Dr. Waymack and Dr. Nartey, whose guidance and insight kept us focused and grounded throughout the project. Reflecting on this experience, we feel we have learned so much about pharmacovigilance and the FDA alongside our own personal journeys and are extremely thankful of both the Capstone Program and the CBHDS for working with us on this project.
Project Team Members:
- Edan McDonald, B.S. in Computation Modeling and Data Analytics (Minor: Computer Science, Chinese, Statistics), May 2025
- Shweatha Rameshkumar, B.S. in Computational Modeling and Data Analytics, Statistics, and Computational Neuroscience (Minor: Computer Science, Mathematics, Adaptive Brain and Behavior), May 2025
- Yimmi Tran, B.S. in Computational Modeling and Data Analytics Cryptography and Cybersecurity concentration (Minor: Computer Science, Mathematics), May 2025.
- Jackson Walsh, M.P.H with a concentration in Infectious Disease, B.S. in Computational Modeling and Data Analytics with a concentration in Biological Science and B.S. in Public Health, May 2025
CBHDS Sponsors:
2024 Projects
- Project Aim: Exploring adverse reactions linked to Singulair and similar drugs, aiming to identify psychiatric safety signals
- Analytic methods: Bayesian methods, disproportionality analysis, and logistic regression
- Visual analytics: Heat maps, ROC curves, and time-series plots

Team's Story
Our project explored adverse reactions linked to Singulair and similar drugs, aiming to identify psychiatric safety signals and assess FDA regulatory decisions. We analyzed data from the FDA’s Adverse Event Reporting System (FAERS) using logistic regression, Bayesian methods, and disproportionality analysis to detect patterns in adverse events, focusing on psychiatric reactions like depression and suicidal ideation. Visualizations included heatmaps, ROC curves, and time-series plots, revealing notable trends and providing a comprehensive view of the data. The results highlighted significant safety signals for Singulair over its comparable drugs, emphasizing the importance of pharmacovigilance.
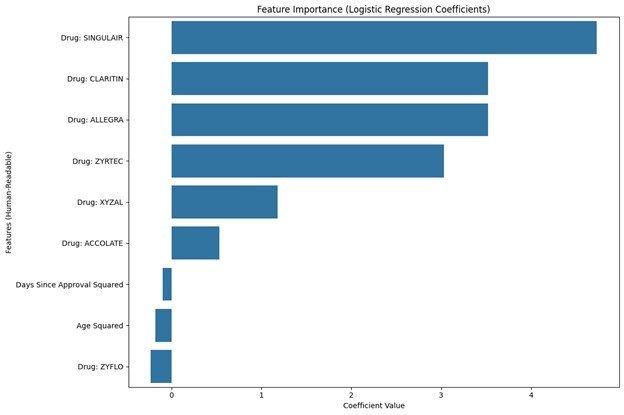
Experiential Narrative
Working on this project was a blend of excitement and challenges. At first, the vast dataset and the complexity of pharmacovigilance analysis were intimidating. Diving into tools like logistic regression, Bayesian analysis, and FAERS data cleaning required us to step out of our comfort zones. Thankfully, teamwork and shared learning helped us push through. There were moments of frustration—like reconciling discrepancies in the data and fine-tuning our models—but seeing the insights unfold was deeply rewarding.
One of the most memorable aspects was the collaborative troubleshooting. Whether it was refining heatmaps or resolving technical issues, everyone contributed their expertise and supported each other. Looking back, this project not only deepened our understanding of adverse drug reactions but also strengthened our problem-solving skills. It was a unique journey, blending data science with public health, and we’re proud of the impact our analysis could have on drug safety assessments.
Project Team Members:
- Hyunjoon Choi, B.S. in Computational Modeling and Data Analytics (Minor: Mathematics), December 2024
- Gabriel Dell, B.S. in Computational Modeling and Data Analytics (Minor: Mathematics), May 2025
- Katelyn Landgraf, B.S. in Computational Modeling and Data Analytics (Minor: Mathematics), May 2025
- Coby Nguonly, B.S. in Computational Modeling and Data Analytics (Minor: Computer Science), May 2025
CBHDS Sponsors:
- Guiding Question: What are the genes responsible for the defense of the Bacterial Leaf Streak pathogen?
- Analytic methods: RNA-sequencing, normalization, pairwise comparisons
- Visual analytics: Heat maps and volcano plots.
Team's Story
Our project aimed to examine the specific genes responsible for the defense of a tobacco plant with an R (defense) gene when signaled by an effector gene in the pathogen. Our clients and their team set up an experiment with three groups (Baseline, T, and NT) to single out specific genes expressed in the T group. We used RNA-Sequencing to process/clean the mRNA samples and align them to the tobacco plant reference genome and count the occurrences of all the genes in each sample. Then through statistical analyses (including normalization and pairwise comparisons), we found the significantly differentially expressed genes and created heatmaps and volcano plots to visualize differences in gene expression levels between treatments.
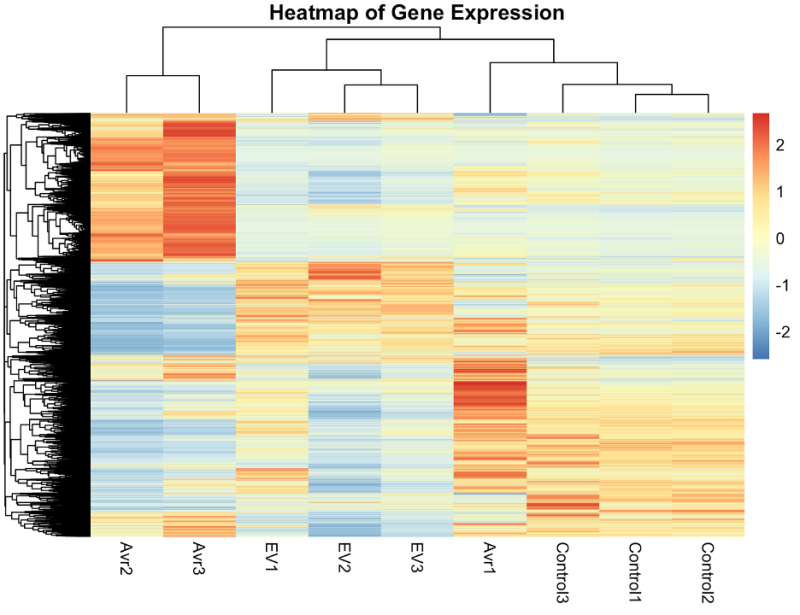
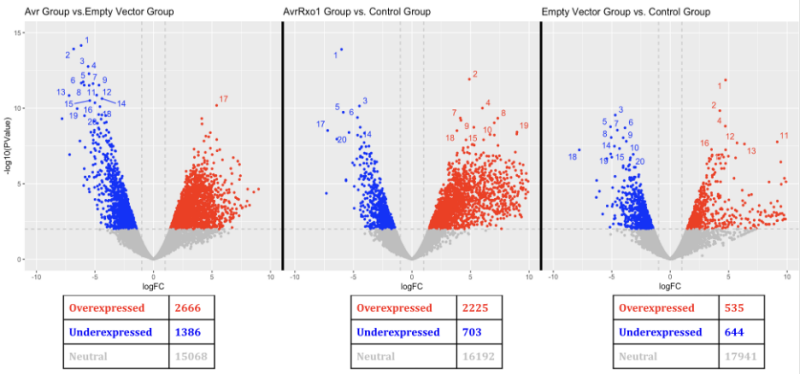
Experiential Narrative
At first our team felt fairly overwhelmed by the amount of new information we would have to learn. None of us had any experience in biology and had never taken any classes and so we went in expecting this project to be constant reading and researching. Luckily though, our sponsor Dr. Zhang helped us a lot in teaching us the basics of what we would need to know for our project, giving us presentations, talking us through our many questions and giving us other resources such as articles to read through to get a better understanding.
Learning the tools was also difficult, getting accustomed to the LinuxOS in the VT ARC system, how to write bash scripts and how high-performance computing works was also hard but fun to learn. We gained a lot of knowledge and experience in the world of Bioinformatics and even though we probably won’t be going into the field directly, it did give us a lot of confidence in knowing that we were able to learn, understand, and produce results in a field we previously thought was so alien and confusing. Again, we are super thankful for Dr. Zhang, and Tanner Barbour for helping us out through the project, we couldn’t have done it without y'all.
Project Team Members:
- Alex Vidal, B.S. in Computational Modeling and Data Analytics, May 2024
- Yelebe Desta, B.S. in Computational Modeling and Data Analytics, May 2024
- Ian Sekelesky, B.S. in Computational Modeling and Data Analytics, May 2024
CBHDS Sponsors:
Previous Projects
- Guiding Question: Is there a pattern to the spread of Salmonella?
- Analytic methods: Comparative genomic analysis
- Visual analytics: Robust phylogenetic trees
Team's Story
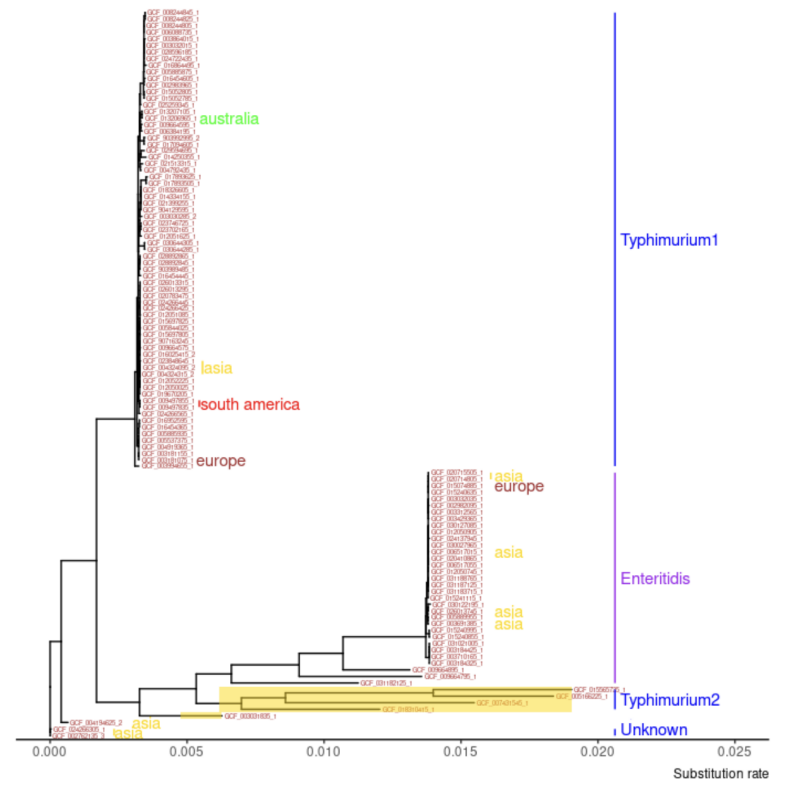
The main objectives of our project were to retrieve genomic data for over 100 strains of Salmonella enterica and use bioinformatic tools to extract and compare their gene families. We performed comparative genomic analysis on these genomes to identify genetic similarities and differences to determine evolutionary relationships. All of this work led to the creation of a robust phylogenetic tree visualization that was able to show common ancestors of the genomes as well as geographic transmission patterns. This tree gives insight into evolutionary trends of Salmonella and gives a clear visualization of how our collected genomes were related to one another.
Our team was initially hesitant in approaching this project when reading its description and seeing new research methods that we had never heard of. However, in our first Zoom meeting with the CBHDS we were walked through an overview of the project and learned many of the biology-related terms related to the project. We immediately became interested in the work that we had to do because we knew the process and the finish line.
Experiential Narrative
It was frustrating when we failed at using the bioinformatic tools, but it was rewarding when we were able to run them correctly with help from our Dr. Zhang. Finally, seeing our results and the final phylogenetic tree gave a unique insight into the concept of evolution, as we were able to see specifically how all our collected strains of Salmonella were related to each other, and how certain strains evolved over time to create new, unique strains.
Working on a bioinformatics project was a first for the CMDA Capstone program, and we are very glad we had the chance to be the trailblazers for this unique project and experience.
Project Team Members:
- Judson Powers, B.S. in Computational Modeling and Data Analytics, May 2024
- Nicholas Emig, B.S. in Computational Modeling and Data Analytics, May 2024
- Siddarth Ravikanti, B.S. in Computational Modeling and Data Analytics, May 2024
CBHDS Sponsors:
- Guiding Question: Can we better understand the dominating preference for ultra-processed foods over minimally processed foods in America?
- Analytic methods: Linear mixed effects modeling
Team's Story
Working as a group was tough at times but with help from one another along with the support from Dr. Ahrens and Ms. Lozano we were able to work through it. The experience was something that we never had before in a normal classroom setting. The project allowed us to get a taste of what it would be like in the real world in terms of large-scale projects. In the end, although there was uncertainty at times, we were proud of our final product and the hard work made it that much sweeter.
Project Team Members:
- Jacob Parker, B.S. in Computational Modeling and Data Analytics
- Sabrina Hart, B.S. in Computational Modeling and Data Analytics
- Aaron Ni, B.S. in Computer Science and B.S. in Computational Modeling and Data Analytics
- Anish Monokonda, B.S. in Computational Modeling and Data Analytics
CBHDS Sponsors:
- Guiding Questions:
Can we find demographic and food perception factors that predict overall food preference across respondents of a national survey?
Additionally, do people prefer ultra-processed foods over minimally processed foods overall? - Analytic methods: Linear mixed effects modeling
- This team was recognized by Mr. Brian Sanchez and Ms. Nancy Schuessler with an honorary sponsorship of their project.
Team's Story
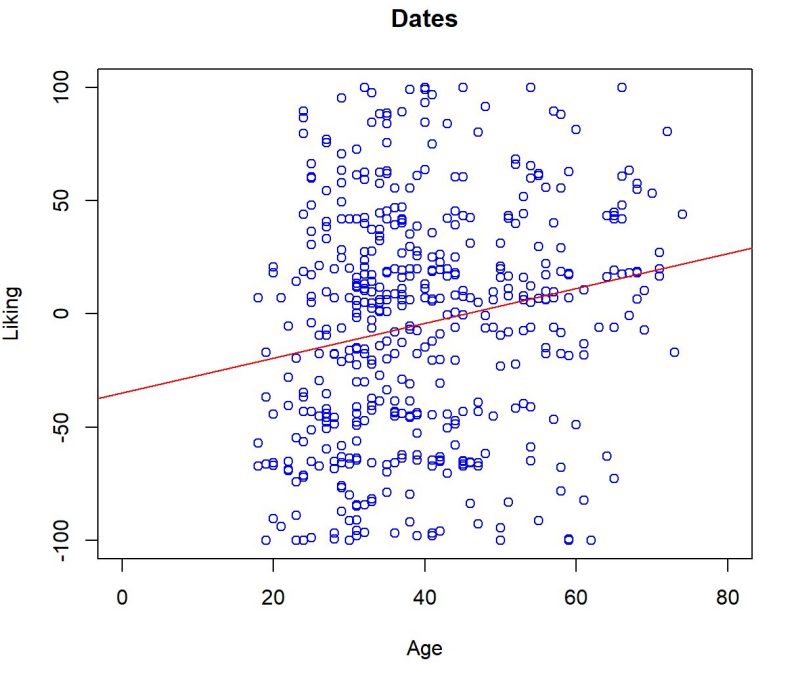
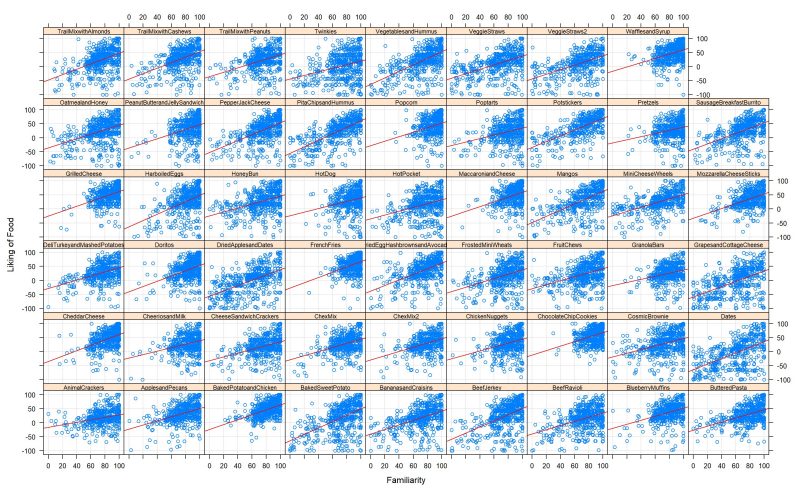
Being a part of a new study, Big Byte Analytics had the opportunity to tackle the poor diet issue caused by ultra-processed foods (UPFs) in the United States. We were interested in finding what factors lead to increased UPF intake using survey data and nutrition information data. Over the course of the project, we have faced significant delays in data collection and we unfortunately were unable to conduct analysis on the true nutrition information of the foods featured in the survey. We hope that future teams can use this data along with our cleaned data and results to answer our original research question.
Regardless of our setbacks, we have found some meaningful results that we believe will serve as a great first step towards finding a solution to the poor diet problem in the United States. We have found that an individual’s age and perception of food (i.e. how healthy they think the food is, perceived calorie count, etc.) has significant influence on food preference. We have also found that most people do not have a particular preference between UPFs and minimally-processed foods and concluded that high UPF intake could also be strongly influenced by factors such as low cost, accessibility, advertising, and lack of health/food knowledge.
Project Team Members:
- Laura Nury, B.S. in Computational Modeling and Data Analytics (CryptoCyber Option, Minor in Mathematics), May 2023
- Rithvik Guntor, B.S. in Computational Modeling and Data Analytics (Minor: Mathematics and Computer Science), December 2022
- Renny Adjei, B.S. in Computational Modeling and Data Analytics (Minor: Statistics and Mathematics), May 2023
CBHDS Sponsors:
- Guiding Question: Can we build on the work done by team Diet Code and expand the predictive modeling of cleaned NHANES data to additional metabolic outcomes?
- Analytic methods: Multiple imputation through chained equations, stepwise linear and logistic regression, random forests, neural networks.
- This team was recognized by Mr. & Mrs. Mark and Nancy Scheffel with an honorary sponsorship of their project.
Team's Story
Due to the scope of organizing and cleaning the NHANES data, team diet Code from Spring 2021 was limited in their ability to analyze their data and model diabetes. Building on that work, team Diet Science used the cleaned NHANES data to predict the presence of hypertension and obesity, and to model the amount of LDL cholesterol, a known risk factor for a number of metabolic diseases.
The team first resolved the problem of missing data by sneering that the missingness was at random and then using multiple imputation by chained equations. After imputation, they modeled their outcomes using a combination of stepwise regression, random forests, and neural networks. Of note, the team was able to greatly improve the performance of their initial neural networks by iteratively expanding and refining those models, a process requiring much determination and skill.
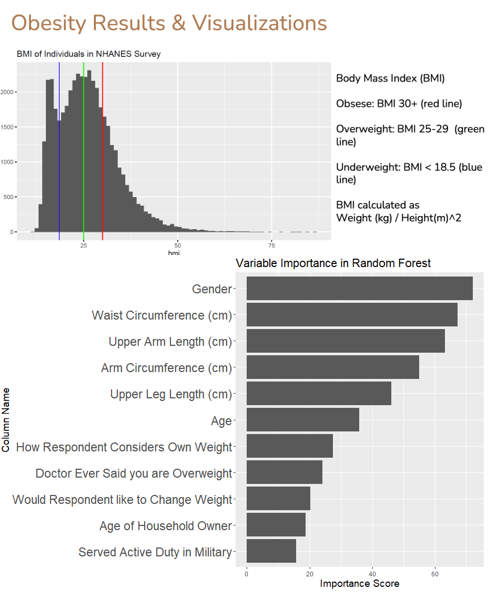
Project Team Members:
- Colin Brant, B.S. in Computational Modeling and Data Analytics, May 2022
- Thomas Stapor, B.S. in Computational Modeling and Data Analytics, May 2022
- Visvas Kaja, B.S. in Computational Modeling and Data Analytics, May 2022
CBHDS Sponsors:
- Ian Crandell
- Alexandra Hanlon
- Guiding Question: Can we harmonize and compile several years of NHANES data and use it to predict diabetes?
- Approaches: In-depth study of multiple years of messy survey data, manual harmonization and data manipulation
- Analytic methods: Stepwise logistic regression
Team's Story
This project was somewhat different from traditional CMDA projects as its main objective was to tackle a problem which, while common, is not often discussed in class. Typically, a student begins their analytic process with a clean data set to which they can immediately apply whatever summary or analytic method they choose. This is rare in practice, which more commonly begins with a very disorganized collection of data that needs to be transformed into a usable form. For instance, some variable names correspond to different questions in different years, and the only way to resolve these discrepancies is with meticulous study.
After this impressive step, the data was still plagued with issues such as missing data, multicollinearity, and potential sampling bias. The team resolved these via complete case analysis and stepwise logistic regression, and were able to predict diabetes in the sample with a high level of accuracy. This project taught the team the value of careful study and exposed them to an often underappreciated dimension of data analysis.
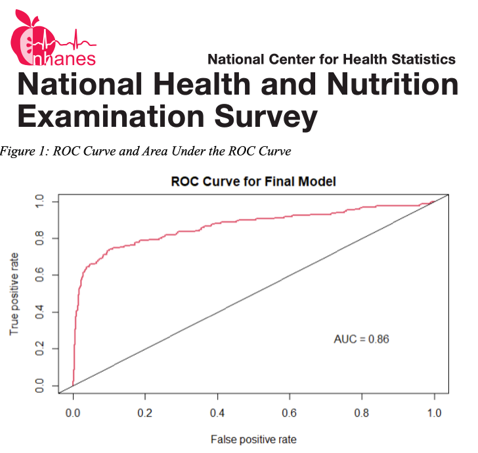
Project Team Members:
- Lauren Bradley, B.S. in Computational Modeling and Data Analytics, May 2022
- Evan Briscoe, B.S. in Computational Modeling and Data Analytics, December 2021
- Jake Lavitt, B.S. in Computational Modeling and Data Analytics, December 2021
CBHDS Sponsors:
- Ian Crandell
- Xin Xing
- Alexandra Hanlon
- Objectives: To study the impact of COVID-19 on the Virginia real estate market and explore inter- and intra-county mobility trends.
- Guiding Quesion: How did COVID-19 impact home values and mobility trends throughout Virginia?
- Analytic methods: Linear Regression and Spatial Regression
- Visual Analytics: An interactive application with Choropleth Maps and Time Series Plots
Team's Story
This project started off very open ended, and we were responsible for coming up with our own research question. With the help of our client and coach, we decided to focus on home values, COVID-19 cases, and mobility trends in Virginia at the county level. From there, our group worked together to produce a final product, while meeting with our client and coach on a weekly basis to brainstorm ideas and go over our progress.
Although we had experience working with data in the past, we ran into some novel concepts such as spatial regression and creating dashboards that took some time to get accustomed to. This project was one of our first experiences working on a team for an extended period of time, and it helped us better understand the importance of team dynamics and time management. Overall, working with CBHDS was a great experience, and we learned a lot throughout the process.
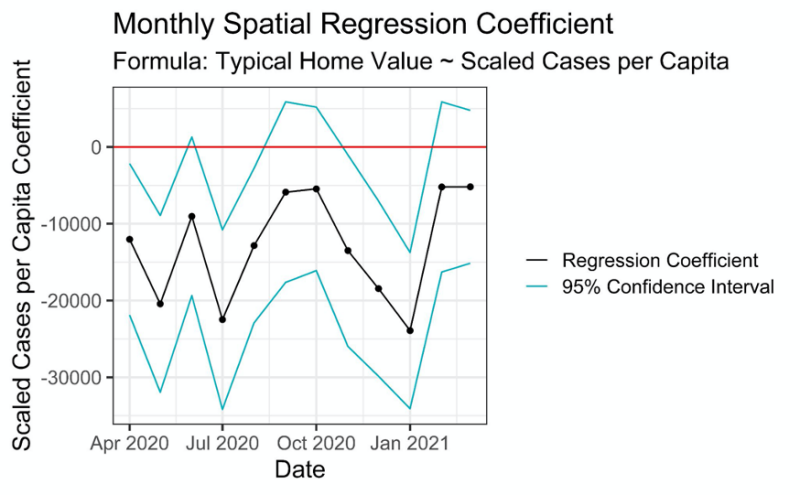
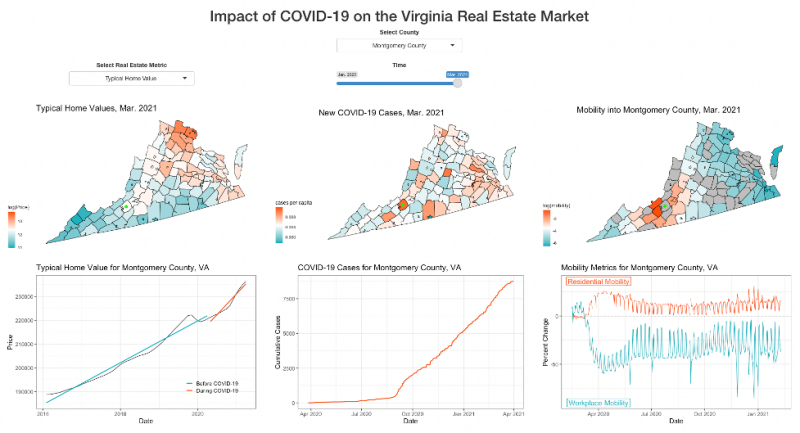
Project Team Members:
- Yohannes Afework, B.S. in Computational Modeling and Data Analytics, May 2021
- Devon Lee, B.S. in Computational Modeling and Data Analytics, May 2021
- Jaffar Shaik, B.S. in Computational Modeling and Data Analytics, May 2021
- Naod Teklie, B.S. in Computational Modeling and Data Analytics, May 2021
CBHDS Sponsors:
- Ian Crandell
- Alicia Lozano
- Alexandra Hanlon
- Guiding Question: How has COVID-mandated social distancing affected United States mental health outcomes at the State level?
- Visual analytics: An interactive application to demonstrate the change in Mental Health Scores week by week during the pandemic

Team's Story
Over the course of the semester, Team New Horizons has been working with CBHDS to study the effects of social distancing on mental health for their Computational Modeling and Data Analytics (CMDA) capstone project. The team consists of Jeff Straw, Demory Williamson, Xumanning Luo, and Bella Marku, all of whom are seniors in CMDA. Their primary motivation for choosing the topic was the opportunity to learn more about how the pandemic has affected people emotionally, not just physically.
The project incorporated both mobility data from Google and mental health data from the CDC to determine how mental health symptoms changed in relation to the amount of time spent at home, as compared to a baseline from before the pandemic began. To display their results, the team developed an interactive web dashboard where users can click on states to obtain that state’s specific mental health results since the start of the pandemic. The biggest challenge the team faced was finding publicly available data, especially since the pandemic is still ongoing, but the datasets they incorporated allowed them to create a comprehensive dashboard that provides significant information for its users.
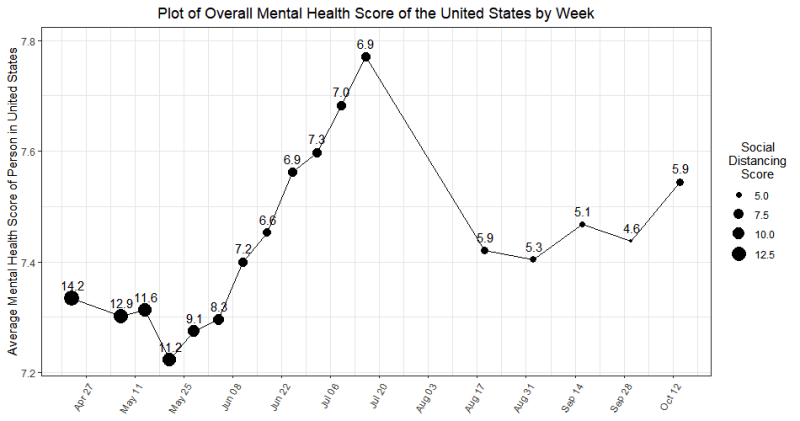
Project Team Members:
- Xumanning Luo, B.S. in Computational Modeling and Data Analytics, May 2021
- Bella Marku, B.S. in Computational Modeling and Data Analytics, May 2021
- Jeff Straw, B.S. in Computational Modeling and Data Analytics, May 2021
- Demory Williamson, B.S. in Computational Modeling and Data Analytics, May 2021
CBHDS Sponsors:
- Ian Crandell
- Kevin McKee
- Alexandra Hanlon
- Guiding Question: How do remediation measures affect species spread?
- Analytic methods: Generalized linear models and classification trees
- Visual analytics: An interactive application to demonstrate the effect of various parameters and conditions
Team's Story
Our CMDA capstone project began with the unexpected challenge of working around COVID-19, but ultimately the project provided us with an entirely unique and informative experience. Extensive collaboration on such a large project was difficult over solely the internet. However, with the guidance of Dr. Alexandra Hanlon and Jennifer West, we became familiar with working efficiently online. Dr. Hanlon and Jennifer were extremely helpful in advising us and providing us with the resources to succeed. We couldn’t have done this without them!
For as unusual as this semester has been, it went by extremely fast. Our entire team feels like we just became acquainted with the CBHDS team. This seemingly short experience has provided us with valuable skills in not only data science, but more so in the importance of teamwork and communication. It was exciting to be able to apply what we have learned in the classroom to a real world problem, and a privilege to do so under the guidance of our mentors.

Project Team Members:
- Evan Mitchell, B.S. in Computational Modeling and Data Analytics, May 2021
- Colton Mumley, B.S. in Computational Modeling and Data Analytics, December 2020
- Akshay Patel, B.S. in Computational Modeling and Data Analytics, May 2021
CBHDS Sponsors:
- Jennifer West
- Alexandra Hanlon



